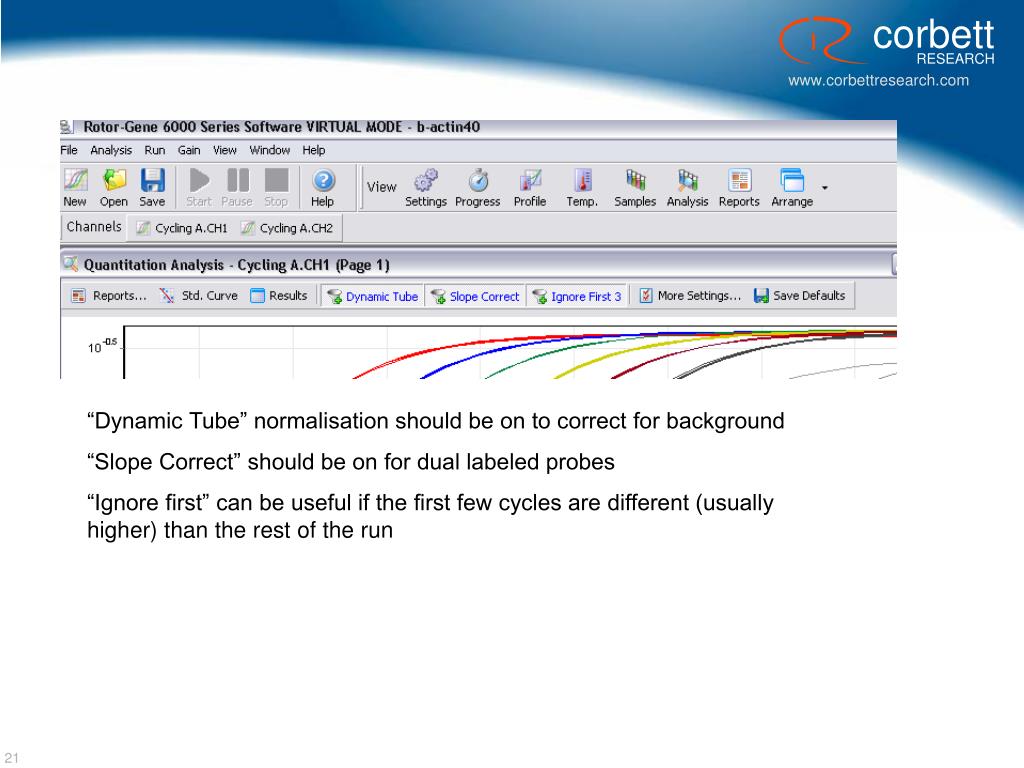
Download Rotor Gene 6000 Software
REST 2008 is astandalone software package for analyzing gene expression usingreal-time amplification data. The software addresses issues surroundingthe measurement of uncertainty in expression ratios by introducingrandomization and bootstrapping techniques.
New confidence intervalsfor expression levels also allow measurement of not only thestatistical significance of deviations but also their likely magnitude,even in the presence of outliers. Whisker box plots provide a visualrepresentation of variation for each gene, highlighting potentialissues such as distribution skew. REST 2008 builds on its predecessorREST 2005 with significant improvements to randomization algorithms.This new revision introduces alternative data inputs such as singlesample efficiency and amplification take-off point, alleviating theneed to set amplification plot thresholds. REST2005 is a new standalone software tool to estimate up anddown-regulation for gene expression studies.
Thesoftware addresses issues surrounding the measurement of uncertaintyforexpression ratios, by using randomisation andbootstrapping techniques. By increasing the number of iterations from 2,000to 50,000 in this version hypothesis tests achieve a level ofconsistencyon par with traditional statistical tests. Newconfidenceintervals for expression levels also allow scientists to measurenotonlythe statistical significance of deviations, but also their likelymagnitude, even in the presence of outliers.
Graphicaloutput of the data via a whisker box-plots provide a visual representationof variation for each gene that highlights potential issues such asdistribution skew. =download here:Newfeatures in REST-384:- upto 15 genes c an be analysed-up to 20 replicated per group- more referencegenes can be chosen- calculation of an geometric mean of the chosen RGs = RG Index- optimal for high throughput 96- and 384-well plate qPCRapplications- efficiency calculation viadilution row-manual efficiency input- data output in agraph with error bars-error estimation of the calculated ratio using a Taylor's series- bugs removed in randomisation testREST-RGbetasoftwareversion 3 August 2006 =download here:New features in REST-RG.
up to 15 genes c anbe analysed- up to 20 replicated per group- more referencegenes can be chosen- calculation of an geometric mean of the chosen RGs = RG Index-optimalfor applications- direct import ofRotor-Gene take off points (TOP) via copy-and-paste- directimport ofsingle-run qPCR amplificationefficiencies via copy-and-paste- manual efficiency input- data output in a graph with error bars-error estimation of the calculated ratio using a Taylor's series- bugsremoved in randomisation test. Real-timequantitativepolymerase-chain-reaction (qPCR) is a standard technique in mostlaboratories used for various applications in basic research. Analysisof qPCR data is a crucial part of the entire experiment, which has ledto the development of a plethora of methods. The released tools eithercover specific parts of the workflow or provide complete analysissolutions.
Here, we surveyed 27 open-access software packages and toolsfor the analysis of qPCR data. The survey includes 8 Microsoft Windows,5 web-based, 9 R-based and 5 tools from other platforms. Reviewedpackages and tools support the analysis of different qPCR applications,such as RNA quantification, DNA methylation, genotyping, identificationof copy number variations, and digital PCR. We report an overview ofthe functionality, features and specific requirements of the individualsoftware tools, such as data exchange formats, availability of agraphical user interface, included procedures for graphical datapresentation, and offered statistical methods. In addition, we providean overview about quantification strategies, and report variousapplications of qPCR. Our comprehensive survey showed that most toolsuse their own file format and only a fraction of the currently existingtools support the standardized data exchange format RDML. To allow amore streamlined and comparable analysis of qPCR data, more vendors andtools need to adapt the standardized format to encourage the exchangeof data between instrument software, analysis tools, and researchers.
IntroductionGene expression analysis isbecoming increasingly important in biological research and clinicaldecisionmaking, with real-time quantitative PCR becoming the method of choicefor expression profiling of selected genes. Maturation of chemistryand hardware has made the practical performance of real-timequantitativePCR measurements feasible for most laboratories. However, accurate andstraightforward mathematical and statistical analysis of the raw data(cycle threshold values) as well as the management of growing data setshave become the major hurdles in gene expression analyses. Since thesoftwareprovided along with the different detection systems does not provide anadequate solution for these issues, we developed qBase, a free softwareprogram for the management and automated analysis of real-timequantitativePCR data. What is qBase?
Corbett Rotor Gene 6000 Manual Pdf
QBaseis a collection of macrosfor Microsoft Excel (currently only Windows version) for the managementand automated analysis of real-time quantitative PCR data. The programemploys a delta-Ct relative quantification model with PCR efficiencycorrection and multiple reference gene normalization. The qBase Browserallows data storage and annotation by hierarchically organizingreal-time PCR runs into projects experiments runs.
It iscompatible with the export files from many currently available PCRinstrument softwares and provides easy access to all your data, bothraw and processed. The qBase Analyzer contains an easy run (plate)editor, performs qualitycontrol and inter-plate calibration, converts Ct values into normalizedand rescaled quantities with proper error propagation, and displaysresults both tabulated and in graphs. The program can handle anunlimitednumber of samples, genes and replicates, and allows data from multipleruns to be processed together (preceded by an inter-run calibrationif required). The possibility to use up to 5 reference genes allowsreliable and robust normalization of gene expression levels. QBaseallowseasy exchange of data between users, and exports tabulated data forfurtherstatistical analyses using other dedicated software.
Real-time PCR is being usedincreasingly as the method of choice for mRNA quantification, allowingrapid analysis of gene expression from low quantities of startingtemplate. Despite a wide range of approaches, the same principlesunderlie all data analysis, with standard approaches broadly classiffedas either absolute or relative. In this study we use a variety ofabsolute and relative approaches of data analysis to investigatenocturnal c-fos expression in wild-type and retinally degenerate mice.In addition, we apply a simple algorithm to calculate the amplifcationeffciency of every sample from its amplifcation profle. We confrm thatnocturnal c-fos expression in the rodent eye originates from thephotoreceptor layer, with around a 5-fold reduction in nocturnal c-fosexpression in mice lacking rods and cones.
Furthermore,weillustrate that differences in the results obtained from absolute andrelative approaches are underpinned by differences in the calculatedPCR effciency. By calculating the amplifcation effciency from thesamples under analysis, comparable results may be obtained without theneed for standard curves.
We have automated this method to provide ameans of streamlining the real-time PCR process, enabling analysis ofexperimental samples based upon their own reaction kinetics rather thanthoseof artificial standards. The GeneExpression Macro is a simple tool forcalculating relative expression values from real-time PCR datagenerated by the iCycler iQ or MyiQ systems. Bio-Rad developed the GeneExpression Macro as a Microsoft Excel workbook containing specializeddata analysis functions.
Download Rotor Gene 6000 Software For Windows 10
Use this macro to save valuable time byemploying standard methods of relative gene expression analysis inpre-designed, easy-to-use Excel spreadsheets.The macro workbooks provided here have been tested with Excel 2000 andExcel 2003, running on a Windows 2000 or XP platform. These files have notbeen tested usingany of the following computing platforms:. Windows 98 or Windows ME. Excel on the Macintosh.
Any other workbook or spreadsheet programs. Quantitativereal-time PCR represents a highly sensitive andpowerful technique for the quantitation of nucleic acids.
Ithas a tremendous potential for the high-throughputanalysis of gene expression in research and routine diagnostics.However, the major hurdle is not the practical performanceof the experiments themselves but rather the efficient evaluation andthe mathematical and statistical analysis of the enormous amountof data gained by this technology, as thesefunctions are not included in the software provided by themanufacturers of the detectionsystems. In this work, wefocus on the mathematical evaluation and analysis of the data generatedby quantitative real-time PCR, the calculation of thefinal results, the propagation of experimental variation of themeasured values to the final results, and the statistical analysis.We developed a Microsoft® Excel®-based software applicationcoded in Visual Basic for Applications, called Q-Gene, whichaddresses these points. Q-Gene manages and expedites the planning,performance, and evaluation of quantitative real-time PCR e xperiments,as well as the mathematical and statistical analysis, storage, andgraphical presentationof the data. The Q-Gene software application is a tool to copewith complex quantitative real-time PCR experiments at ahigh-throughput scale and considerably expedites and rationalizes theexperimentalsetup, data analysis, and data management while ensuring highestreproducibility. Quantitativereal-time PCR representsa highlysensitive and powerful technique for thequantitation of nucleic acids.
It has a tremendous potentialfor the high-throughput analysisof gene expression in research androutine diagnostics. However, the major hurdle is not thepractical performance of the experiments themselvesbut rather the efficient evaluation and the mathematical and statistical analysis ofthe enormous amount of data gainedby this technology, asthese functions are not included inthe software provided bythe manufacturers ofthe detectionsystems.
In this work, we focus on the mathematicalevaluation and analysis of the data generated by quantitative real-time PCR,the calculation of the final results, the propagation of experimental variation of the measuredvaluesto the final results, and the statisticalanalysis. We developed a Microsoft Excel-based software application coded in Visual Basic forApplications, called Q-Gene, whichaddresses these points.Q-Gene manages and expeditesthe planning, performance, and evaluation and quantitative real-timePCR experiments, as well as the mathematical and statisticalanalysis, storage, and graphical presentation of the data.
The Q-Gene softwareapplication is a tool to cope with complexquantitative real-timePCR experiments at ahigh-throughput scale and considerably expedites andrationalizes the experimental setup, data analysis, and data management while ensuring highestreproducibility.Erratum for: MullerPY, Janovjak H, MiserezAR, Dobbie Z.Processing of gene expressiondata generated by quantitative real-timeRT-PCR.Biotechniques. 2002 32(6): 1372-1378. In Table 1,the values in the column 'NormalizedExpression' need to be replaced by the following ones (top to bottom):2.30E-03, 2.63E-03, 3.92E-03, 2.95E-03, 4.95E-04, 16.79.
Additionally,the values in the column 'Mean NormalizedExpression' need to be replaced by 2.87E-03, 3.26E-04, 11.35. Thedifference between the two calculation procedures accordingto Table 2, Equation 2 and 3, respectively, amounts to 2.8%.Furthermore, the corresponding values in the discussion section need tobereplaced.In all Equations of Table 2, the indices 'target' and 'ref' ofall variables need to be swapped. In Equation 6, a plus sign needs tobe added between the two brackets under the square-root.
TheseEquations have also been corrected in all Q-Gene software files.It is important that you no longer use any former versions of theQ-Gene software files because these files yield wrong results! Itis intended to publish the erratum.Q-Gene: processingquantitative real-time RT–PCRdataPerikles SimonSection for Neurobiology of the Eye, University Eye Hospital Tuebingen,Calwerstr. 7/1, 72076 Tuebingen, Germany.
Summary: Q-Geneis an applicationfor the processing ofquantitative real-timeRT–PCR data. It offers the user the possibility to freely choosebetween two principally different procedures to calculate normalizedgene expressions as eithermeans of Normalized Expressions or Mean Normalized Expressions. In thiscontribution it will be shown that the calculation of Mean NormalizedExpressionshas to be used for processing simplex PCR data, while multiplex PCRdatashould preferably be processed by calculating Normalized Expressions.Thetwo procedures, which are currently in widespread use and regarded asmoreor less equivalent alternatives, should therefore specifically beappliedaccording to the quantification procedure used. Quantitativereal-time PCR is an important high throughput method in biomedicalsciences. However, existing software has limitations in handling bothrelative and absolute quantification. We designed qPCR-DAMS(Quantitative PCR Data Analysis and Management System), a database toolbased on Access 2003, to deal with such shortcomings by the addition ofintegrated mathematical procedures.
QPCR-DAMA allows a user chooseamong four methods for data processing within a single softwarepackage: (I) Ratio relative quantification, (II) Absolute level, (III)Normalized absolute expression, and (IV) Ratio absolute quantification.qPCR-DAMS also provides a tool for multiple reference genenormalization. QPCR-DAMS has three quality control steps and a datadisplay system to monitor data variation. In summary, qPCR-DAMS is ahandy tool for real-time PCR users.
LinRegPCR is a programfor the analysis of quantitative RT-PCR (qPCR)data resulting from monitoring the PCR reaction with SYBR green orsimilar fluorescent dyes. The program determines a baselinefluorescence and does a baseline subtraction. Then aWindow-of-Linearity is set and PCR efficiencies per sample arecalculated.
With the mean PCR efficiency per amplicon, the Ct value persample and the fluorescence threshold set to determnine the Ct, thestarting concentration per sample, expressed in arbitrary fluorescenceunits, is calculated = See below:. Ramakers et al., NeuroSci Lett 2003;. Ruijteret al., Nucleic Acids Research 2009.Assumption-freeanalysis of quantitative real-time PCR data. Quantificationof mRNAs using real-time polymerase chainreaction (PCR) by monitoring the productformation with the fluorescent dyeSYBR Green I is being extensively used inneurosciences, developmental biology,and medical diagnostics.Most PCR data analysis procedures assume that the PCR efficiency for the ampliconof interest is constant or even, in the case of the comparative C(t) method,equal to 2. The latter method already leads to a 4-folderror when thePCR efficiencies vary over just a 0.04 range.
PCR efficiencies ofamplicons areusually calculated from standard curves based on either known RNAinputs or ondilutionseries of a reference cDNA sample. In this paper we showthat the firstapproach can lead to PCR efficiencies that vary over a 0.2 range,whereas thesecond approach may beoff by 0.26. Therefore, we propose linear regression on the Log(fluorescence) percycle number data as an assumption-free method to calculate startingconcentrations of mRNAs and PCR efficiencies for eachsample.ThenewLinRegPCR version of the program (with an updated manual) can bedownloaded =. Amplificationefficiency: linking baseline and bias in the analysis of quantitativePCR dataJ.
Ramakers2, W. Hoogaars1, Y.Karlen3, O. Van den Hoff1 and A. Dear LinRegPCR user,RDML wasdeveloped as a standard for export, exchange, and storage ofquantitative PCR data and is supported by several large qPCR systemsuppliers as well as by data analysis software like qbase-plus.LinRegPCR now forms a link between your qPCR system and suchstatistical analysis software. LinRegPCR can handle RDML versions 1.0and 1.1, as well as RDML files in which floating point values arewritten with decimals points and decimal commas. LinRegPCR will writethe analysis results to an RDML file, version 1.1, with decimal pointstomaintain compatibilty with the current RDML specification.The RDML input option is the mainaddition to LinRegPCR that was implemented in 2012.
There were alsoseveral qPCR systems added to the list of input formats from Excelfiles. For other minor changes in the program, please have a look atthe recent updates listed on the LinRegPCR website.On our site you will also find a linkto a recent paper (Ruijter et al., Methods 2012), in which LinRegPCRand other publicly available PCR amplification curve analysis programswere compared. This paper is unique in the field of qPCR because allanalysis methods were applied by their original developers, and thus inthe currently recommended way. The paper was co-authored by thedevelopers of these curve analysis programs and members of the geNormteam, who performed the statistical analysis. The datasets used forthis comparison, and the analysis results, can be downloaded from.I hope you continue to enjoy the useof LinRegPCR.Best wishes for the coming festiveseason and your future scientific endeavours,Jan M Ruijter.
The purpose of thismanuscript is to discuss fluorogenic real-time quantitative polymerasechain reaction (qPCR) inhibition and to introduce/define a novelMicrosoft Excel-based file system which provides a way to detect andavoid inhibition, and enables investigators to consistently designdynamically-sound, truly LOG-linear qPCR reactions very quickly. TheqPCR problems this invention solves are universal to all qPCRreactions, and it performs all necessary qPCR set-up calculations inabout 52 seconds (using a pentium 4 processor) for up to seven qPCRtargets and seventy-two samples at a time – calculations that commonlytake capable investigators days to finish. We have named this customExcel-based file system 'FocusField2- 6GallupqPCRSet-upTool-001'(FF2-6-001 qPCR set-up tool), and are in the process of transforming itinto professional qPCR set-up software to be made available in 2007.The current prototype is already fully functional. Thepurpose of this manuscript is to describe a reliable approach toquantitative real-time polymerase chain reaction (qPCR ) assaydevelopment and project management, which is currently embodied in theExcel 2003-based software program named “PREXCEL-Q” (P-Q) (formerlyknown as “FocusField2-6Gallup-qPCRS et-upTool-001,” “FF2-6-001 qPCRset-up tool” or “Iowa State University Research Foundation ISURFproject #03407”). Since its inception from 1997-2007, the program hasbeen well-received and requested around the world and was recentlyunveiled by its inventor at the 2008 Cambridge Healthtech Institute’sFourth Annual qPCR Conference in San Diego, CA.
P-Q was subsequentlymentioned in a review article by Stephen A. Bustin, an acknowledgedleader in the qPCR field. Due to its success and growing popularity,and the fact that P-Q introduces a unique/defined approach to qPCR, aconcise description of what the program is and what it does has becomeimportant. Sample-related inhibitory problems of the qPCR assay, sampleconcentration limitations, nuclease-treatment, reverse transcription(RT ) and master mix formulations are all addressed by the program,enabling investigators to quickly, consistently and confidently designuninhibited, dynamically-sound, LOG-linear-amplification-capable,high-efficiency-of-amplification reactions for any type of qPCR. Thecurrent version of the program can handle an infinite number of samples. Quantitativereal-time PCR has proven to be an extremely useful technique in lifesciences formany applications. Although a lot of attention has been paid to theoptimizationof the assay conditions, the analysis of the data acquired is oftendone withsoftware tools that do not make optimum use of the information providedby the data.
Particularly, this is the case for high-throughputanalysis, which requires a careful characterization and interpretationof thecomplete data by suitable software. Here we present a software solutionforthe robust, reliable, accurate, and fast evaluation of real-time PCRdata,called SoFAR. The software automatically evaluates the data acquiredwith theLightCycler system. It applies new algorithms for an adaptivebackground correctionof signal trends, the calculation of the effective signal noise, theautomatedidentification of the exponential phases, the adaptive smoothing of therawdata, and the correction of melting curve data.
Finally, it providesinformationregarding the validity of the results obtained. The SoFAR softwareminimizesthe time required for evaluation and increases the accuracy andreliability ofthe results. The software is available upon request.Validationof an algorithm forautomaticquantification of nucleic acidcopy numbers by real-time polymerase chain reactionWilhelm J, Pingoud A, Hahn M.Anal Biochem.
2003 Jun 15;317(2):218-25.Institut fur Biochemie, FB 08, Justus-Liebig-Universitat Giessen,Heinrich-Buff-Ring 58, D-35392 Giessen, Germany. Real-time quantitativepolymerase chain reaction (PCR) with on-line fluorescencedetection has become an important technique not only for determinationof the absoluteor relative copy number of nucleic acids but also for mutationdetection,which is usually done by measuring melting curves. Optimum assayconditionshave been established for a variety of targets and experimental setups,butonly limited attention has been directed to data evaluation andvalidationof the results. In this work, algorithms for the processing ofreal-timePCR data are evaluated for several target sequences (p53, IGF-1, PAI-1,Factor VIIc) and compared to the results obtained by standardprocedures. The algorithms are implemented in software called SoFAR,which allows fully automatic analysis of real-time PCR data obtainedwith a Roche LightCycler instrument.
Thesoftware yields results with considerably increased precision andaccuracyof quantifications. This is achieved mainly by the correction of phaseofthe signal curves.
The melting curve data are corrected for signalchangesnot due to the melting process and are smoothed by fitting cubicsplines.Therefore, sensitivity, resolution, and accuracy of melting curveanalyses are improved. SoFAR ®isa software of biologists and medics for a quantitative analysis andinterpretation of real time PCR measurements. Originally it wasdeveloped by Dr. Jochen Wilhelm for research on a precise quantifyingof tumour suppressor genes and is now distributed and up-dated byMetraLabs ® GmbH exclusively. This softwaremakes a fully automatic analysis and interpretation of measurement datapossible, which are written down by LightCycler ® (RocheDiagnostics ®) or RapidCycler ® (IdahoTechnology). To meet the highest demands of precision and safety in theanalysis, robust algorithms were developed that guarantee reliableresults even with suboptimal data. In combination with a thoughtthrough user friendly surface, real time PCR measureings are easy, fastand precise to analyse.Completeanalysis with just one mouse clickSimplyopen the file whichis to analyse - no other steps are needed.
Therefore one file iscompletelyanalysed with just one mouse click.AccurateresultsSoFARcontrolsautomatically whetherthe criteria for a correct quantitative analysis are obliged. Includedare automatic recognition and evaluation of the exponential phase ofamplification curves as well as the calculated C T values.Curveswhich cannot be analysed correctly are marked.Alwaysbest possible resultsAnefficientnoise-filtering ofthe raw data of amplification and melting curves, makes moreprecise results possible. Independent signal changes from theamplification are automatically recognised and corrected. The automaticcorrection of temperature dependent quenches at melting curves alsoeliminates systematic errors and increases the sensitivity of a meltingcurve analysis.Easydata exportAll results can be printed, saved or exported into otherprogrammes as graphics or in tables. Extensive report functions make anexact documentation of all results easy. Diagrams which can be exportedor copied in publishing quality can be changed and transformed in thelayout from the user.
Availablereal-time PCR cyclers (3).Mic is asmall 48-well qPCR instrument that uses magnetic inductiontechnologyfor rapid real time cycling. The'heart' of the Smart Cycler® System from Cepheid of Sunnyvale,Calif., is the I-CORE™ (Intelligent Cooling/Heating Optical Reaction)module. According to company literature, the I-CORE module incorporatesstate-of-the-art microfluidic and microelectronic design. Each SmartCycler processing block contains 16 independently programmable I-COREmodules, each of which performs four-color, real-time fluorometricdetection.A wide variety of different multiplex or simplex fluorescent tags canused in conjunction with this system, including FAM, TET, ALEXA532, ALEXA 647, TAM,ROX, SYBR Green, Cy3, Texas Red and others. TheGeneXpert® System utilizes real-time polymerase chain reaction(PCR) to amplify and detect target DNA. TheMx™ Family of qPCR Systems from Stratagene offers the highestperformance and flexibility in qPCR Systems at the most affordableprice. The Mx3000P® qPCR System is not just an entry-level QPCRsystem, but is a high-performance, full-featured instrument systemdesigned to accommodate most experimental designs.
The new Mx3005P™qPCR System advances the proven Mx3000P system by offering unmatchedflexibility and capability to support more real-time qPCR applicationsand chemistries. The flexible Mx3005P system will accommodate yourresearch needs now and in the future. Citedin more than3000 publications, the Agilent Mx3005P and Mx3000P QPCR Systems are themost flexible—and reliable—instruments available for gene expressionanalysis, microarray data validation, SNP genotyping, pathogendetection, DNA methylation analysis, and chromatin immuno-precipitationstudies.
Agilent’s qPCR software, MxPro, provides users with anintuitive interface, quick experiment design, powerful data analysisand easy report generation. All of these features and more make qPCRwith the Mx instrument an exciting and dependable user experience.
Citedin more than 3000 publications, the Agilent Mx3005P and Mx3000P QPCRSystems are the most flexible—and reliable—instruments available forgene expression analysis, microarray data validation, SNP genotyping,pathogen detection, DNA methylation analysis, and chromatinimmuno-precipitation studies. Agilent’s qPCR software, MxPro, providesusers with an intuitive interface, quick experiment design, powerfuldata analysis and easy report generation. All of these features andmore make qPCR with the Mx instrument an exciting and dependable userexperience.Mx Pro QPCR software with FDA guideline 21 CFR Part 11 compatiblefeatures. Sacace - SyCycler 96- A new instrument for real-time amplification and melting analysissuitable both for research studies as well as for diagnosticsapplications.